With the constant adaptation and improvement of genetic and gene editing technologies coupled with the refinement of phenotyping approaches, phenomics research is progressing at a remarkable rate. In particular, it is becoming feasible to consider comprehensive, genome-wide, efforts to decipher the genetic networks which underlie the biology of an animal, enabling a more detailed insight into human disease associations.
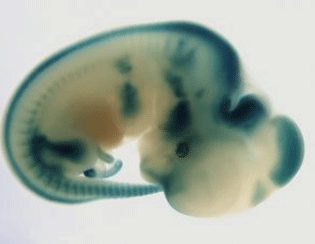
The ultimate challenge is to generate a mutant for every gene in a mammalian genome and to characterise the phenotype of each mutant line. In a newly published Nature Reviews Genetics article, Steve Brown and colleagues explore these advances with a focus on the mouse as a key genetic model. The authors discuss the challenges in developing and applying phenomics tools and outline the impact of different techniques. They conclude by highlighting how wide-ranging analyses of gene function in the mouse allow important insights into human genetics, and in particular the genetic bases of disease. Professor Steve Brown from the MRC Harwell Institute said “ We have tried in this review to give a comprehensive overview of the state of play for mouse high-throughput phenomics and its impact on the study of mammalian gene function and the genetics of disease. It is an exciting time for mouse genetics as developments in generating mouse mutations and analysing the phenotypes of each mutant are bringing us closer to generating a complete functional catalogue of a mammalian genome.” Background Over the years many different methods have been utilised to study genetic mutations in the mouse – including the identification of natural variants in breeding stocks, and the use of radiation and chemicals to generate mutants. These methods offered valuable insights, but were limited as they were not able to target specific loci within the genome and ultimately only allowed a small number of phenotypes and genes to be explored. This has changed over the last couple of decades, and it’s now possible to target specific genes, and a wide variety of mutant alleles for each gene can be produced. The advent of CRISPR-Cas9 has revolutionised this process and allows a fast, economical method for creating targeted mouse mutations. A range of other uses for CRISPR are also in development. In addition, the development of technologies for the archiving and distribution of mutant lines, such as the use of dry ice for the shipment and storage of mouse sperm, enables institutions to easily exchange and share mutant CRISPR-Cas9 lines. “The use of CRISPR technology is further expanding the genetic toolkit by which we can alter the mouse genome and revolutionising our ability to understand the function of genes.” Phenotyping Platforms Along with the exciting developments in our toolkit for modifying the mouse genome, parallel and important advances in phenomics are enabling us to undertake a comprehensive assessment of phenotype for each and every mutant line. Sophisticated phenotyping platforms, in which a range of tests examine different aspects of physiology, biochemistry and development, have been developed. Comprehensive phenomics approaches assessing diverse biological systems will allow an in depth analysis of genetic networks and disease. This is exemplified by the study of pleiotropy. It is becoming clear that this phenomenon, whereby genes have multiple functions, is extremely common. Therefore, if phenotyping approaches are not sufficiently broad and comprehensive these relationships can be missed or misinterpreted. Thus to fully explore pleiotropy, considerable emphasis is being placed on the development of an increasing sophistication of phenotyping platforms encompassing all body systems so that a diverse range of biological mechanisms can be studied. This broad phenotyping pipeline approach therefore offers more comprehensive insights into gene function. The simplest phenotyping methods involve direct cage side observations, documenting dysmorphologies and variations in size. Unusual behaviour and movements can also be identified by experienced researchers and laboratory technicians. On top of these simple, although highly useful, observations, a great variety of other tests have been developed. These include phenotypes relating to exercise, balance, neurology, bone structure, body composition, clinical chemistry, hearing and metabolism. Crucially, these tests have a relatively low impact on individual mice and therefore multiple phenotyping platforms can be used, and repeated, during an animal’s lifetime. Additionally, after death important phenotype information can also be gathered, through histopathology and, for lethal mutants, assessment of morphology at different embryonic stages. “We are at a new frontier for mouse phenomics. Revolutionary advances in our ability to measure and assess diverse systems including physiology, biochemistry and behaviour will provide us with profound insights into how genes impact on biology and disease.” International Mouse Phenotyping Consortium An example of where this new technology and phenotyping practices are implemented is the International Mouse Phenotyping Consortium (IMPC). This consortium was formed in 2011 with the aim of generating a catalogue of mouse gene function through the use of knockout mouse. At the beginning of this project mutants were generated by using tm1b null mutant allele. However the IMPC now uses CRISPR-Cas9 to generate the null mutations by deleting an early critical exon. Homozygous mutants enter a standardised and broad adult phenotyping pipeline. Both male and female mutant mice undergo a range of tests including behavioural, metabolic, neurological, cardiovascular, sensory and musculo-skeletal function. A variety of terminal tests are then undertaken. Characterisation of embryonic lethal homozygotes through an embryonic pipeline also takes place employing high resolution imaging approaches. For embryonic lethal mutants heterozygotes enter the adult phenotyping pipeline and a lacZ reporter allows determination of tissue expression of the disrupted gene. Phenotype data has now been collected for over 5,000 mutant IMPC lines. 90% of the gene-phenotypes described by the IMPC are new, highlighting widespread pleiotropy. Furthermore, there is a strong connection between phenotypes described in mouse knockouts and those associated with human disease genes, with 40% of IMPC lines having phenotypic overlap with their human counterpart. Importantly, IMPC data has also published a number of other key observations, including the variability of embryonic lethality in mutant lines, and the high rate of sexual dimorphism in mutant phenotypes. “The IMPC is rising to the challenge of generating a complete functional catalogue of the mouse genome. Since its inception in 2011, it has made great strides with a third of the genome already analysed. Moreover, many startling and hitherto undiscovered features of the mammalian genome landscape have been revealed.” One of the main challenges in using mice in genetic studies is being able to extrapolate mouse behaviour to humans. New phenotyping tests are beginning to address this issue, with more complex protocols allowing more comparable behavioural data to be collected. Another challenge that is being addressed is age-related phenotypes, with some mutant lines now aged and re-phenotyped. Analysis of all the phenotyping data collected is also a crucial step. These methods have been refined in recent years, with different tests employed to analyse and display results, with linear regression and Bayesian approaches allowing modelling of factors beyond the genotype and phenotype data. New methods to analyse and interpret 3D data are also in development and offer a more robust means of interpreting data, including machine learning techniques. In summary, high-throughput mouse phenomics is an important tool for biomedical sciences, and in the next few years this data will increasingly be used alongside human biology studies. Data from IMPC and other mouse phenomics programmes will allow comprehensive insights into human disease processes, with a substantial impact on our understanding of complex disease, and enormous benefits to the Precision Medicine initiative.