An atomic model of a doublet microtubule from mammalian respiratory cilia provides a framework to understand how these proteins allow cilia function and how mutations could cause disease
Our lungs are kept clear of fluid through the action of waving cilia; small hair-like cellular projections that move in a rhythmic manner. In fact, these motile cilia perform a similar function in a range of processes, including determination of left-right asymmetry during development and driving movement of sperm. Dominic Norris’ group, of the Mammalian Genetics Unit at MRC Harwell, in collaboration with the groups of Alan Brown, Sudipto Roy, and others, have published a study in Cell that combines a structural framework for understanding the molecular machines that drive the characteristic beating motion of cilia with in vivo analyses of a pair of previously unstudied proteins.
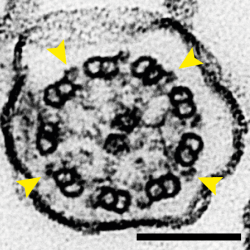
The movement of cilia is driven by dynein motor proteins attached to one doublet microtubule (DMT) attempting to walk along an adjacent DMT. The two DMTs are prevented from sliding along each other, so the action of dynein forces a bending motion instead. Multiple DMTs are arranged in a circle in a structure called the axoneme so that motion generated by dynein molecules on different sides of the circle cause cilia to bend, causing their rhythmic waving. This organisation is visible in the electron micrograph feature image for this story, where nine doublet microtubules are shown to surround a pair of singlet microtubules in a cross-section of a mouse tracheal cilium and yellow arrows point to the bound dyneins.

Atomic model showing PIERCE1 and PIERCE2 spanning the microtubule wall, linking the outer dynein arm docking complex (ODA-DC) to the microtubule inner protein CFAP53
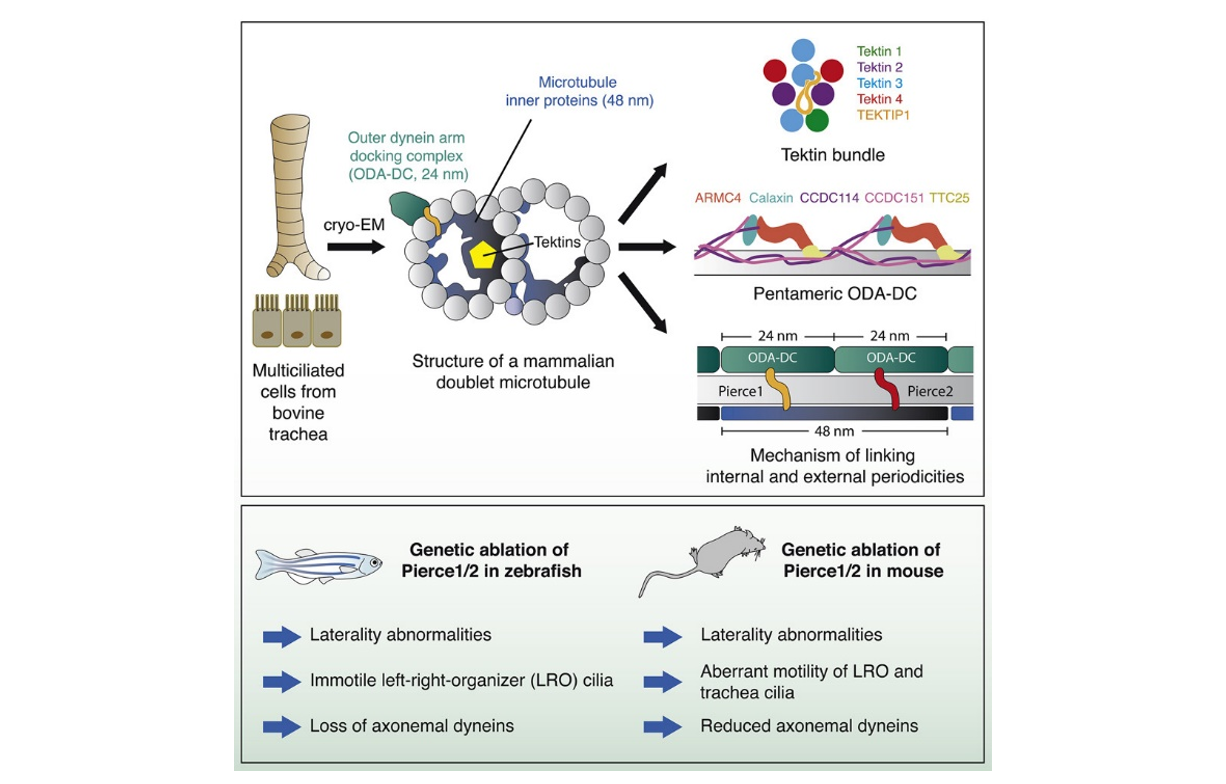
This process depends on interactions between a large number of proteins. Previous work in algae has revealed the precise 3D architecture of these DMT filaments, showing how the presence of docking complexes at regular intervals along the length of the outside of DMTs ensures even spacing of dyneins. It also demonstrated the presence of repeating networks of microtubule inner proteins (MIPs) lining the surface of the tubular DMT structure. However, some of the proteins involved in these complexes are not found in mammals, so further investigation will benefit detailed understanding of the function of the axonemes in our bodies.
To do this, Miao Gui and Jacob Anderson, two of the co-first authors on the paper, and others in Alan Brown’s group at Harvard Medical School, used cryo-electron microscopy (cryo-EM), a technique that allows researchers to determine high resolution images of proteins so that the positions of individual atoms can be revealed and insights into mechanisms of action can be obtained. This allowed the team to build a model of a mammalian DMT and identify 29 MIPs, many of which had not previously been characterised. The structure also demonstrated a type of docking complex for the attachment of dynein that is different to those previously seen in algae.
The model also identified proteins that play important roles connecting the internal network of MIPs within the DMT and the docking complex and dyneins on the outside. The importance of this structural information was demonstrated in two model organisms, mouse and zebrafish. Specifically, two proteins, PIERCE1 and PIERCE2, were investigated using zebrafish (in work led by co-first author Priyanka Anujan working in Sudipto Roy’s group at the Institute of Molecular and Cell Biology in Singapore and Colin Bingle’s group at the University of Sheffield) and mice (in work led by co-first author Hannah Farley in Dominic Norris’ group at MRC Harwell).
Summary of the main findings of the paper
Dominic’s group had first started studying the role of PIERCE1 when working with an IMPC Pierce1 knockout line that showed left/right patterning defects. This drove them to generate mice carrying a null allele of Pierce2 and attempt to cross them to create a double mutant, but high levels of embryonic and pre-weaning lethality were evident in double mutants. Sudipto’s group became interested in pierce1 as it is a target of a key motile cilia transcription factor, FOXJ1. In both of these organisms, the groups found that defects in left-right patterning caused by loss of these proteins could be explained by abnormalities in the beating of cilia that leads to disrupted flow at the left/right organiser in developing embryos and altered asymmetry of gene expression across the left/right organiser. In addition, investigation of tracheal cilia in mice identified a stiffer beating motion, while electron microscopy demonstrated loss of bound dyneins.
This study represents the coming together of different approaches towards understanding disease in order to provide insights from the molecular level through to the level of the whole organism. Thirteen proteins identified in this study are known to be associated with human diseases linked to defects in cilia function and more have disease-like phenotypes in mouse and zebrafish mutants. While the structure provides a framework for understanding how these proteins work together to allow cilia to function normally at the level of molecular mechanisms, the mouse and zebrafish work links genotype to phenotype by directly showing the impact loss of one protein can make on the movement of cilia, which has implications for our understanding and treatment of congenital heart disease and primary ciliary dyskinesia.
This work was supported by UKRI MRC, NIH, the LouLou Foundation, the E. Matilda Ziegler Foundation for the Blind, the Smith Family Foundation, the Pew Charitable Trusts, NHS England, NIHR, The AAIR Charity, and University of Sheffield- and University of Manchester-A*STAR Singapore doctoral studentships.
Gu, M, Farey, H, Anujan, P, Anderson, JR, Maxwell, DW, Whitchurch, JB, Botsch, JJ, Qui, T, Meleppattu, S, Singh, SK, Zhang, Q, Thompson, J, Lucas, JS, Bingle, CD, Norris, DP, Roy, S, Brown, A. De novo identification of mammalian ciliary motility proteins using cryo-EM. Cell. 2021, https://doi.org/10.1016/j.cell.2021.10.007