Recent advances in CRISPR/Cas9 genome editing have made it possible to introduce larger genetic modifications and make more complex genetically altered animal models. However, sequencing to validate that the desired changes have been made can be challenging. The Mary Lyon Centre’s Molecular and Cellular Biology team, led by Lydia Teboul, has piloted how Oxford Nanopore Technology (ONT) long-read sequencing allows fast and robust analysis of genome-editing experiments in their latest article in PLOS Genetics.
Sanger sequencing has been the most common technique used to characterise sites of genetic alterations. However, new methods for the introduction of genome edits have made it possible to create targeted edits spanning several kilobases, which is far longer than a 500–800 bp Sanger sequencing read. To validate edits at this scale, a time-consuming and intricate process is required that involves taking multiple sequencing reads, which are then pieced together in silico. Screens based on this process can produce some false positives and, in animals, allele validation still needs to be repeated in the subsequent generation. To address these issues, the team have assessed the potential for ONT long-read sequencing to replace Sanger sequencing.
ONT sequencing produces long reads, which can cover the entire length of the mutagenized interval, each from a single DNA molecule. Although ONT has a high error rate, the team found that this could be offset with a high sequencing depth, achieved through targeted sequencing to ensure that the region of interest is sequenced many times over so that errors within one read can be ignored. The team assembled a new workflow for the analysis of sequencing data and found that ONT sequencing provides an accurate screen of genetically altered founder animals as well as an efficient tool for definitive validation of the mutant allele in the subsequent generation.
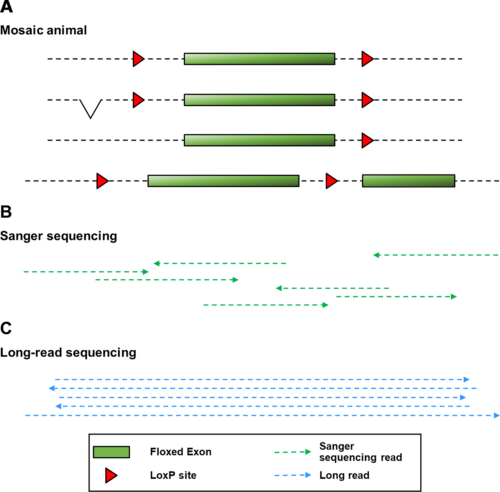
Importantly, they found it to be a relatively simple and user-accessible process, which requires only minimal investment in sequencing equipment. A high maximum number of animals that can be analysed per run makes it cost-effective where the number of new mutants being generated is large enough, but it might be harder to justify the cost of each sequencing run for validation in a smaller number of animals.
The team also compared PCR- and Cas9-based methods for sequencing targeting. While PCR works to amplify the region of interest to be sequenced, nanopore Cas9-targeted sequencing (nCATS) relies on the use of Cas9 to make cuts around the region of interest and the tagging of those cut ends with nanopore sequencing adaptors. PCR has a number of limitations, particularly in the length of product that can be amplified and in the potential for amplification bias, where the presence of a shorter fragment can be preferentially amplified, so it might be that an alternative method to target sequencing efforts might be of benefit. However, the team found that PCR was the more practical option, as nCATS requires a lot of DNA – an amount that is challenging to obtain from an ear biopsy and that might potentially require an animal to be culled, limiting use for founder animals.
Alongside providing a fast and robust system for validation of genome-edited sequences, this methodology also has 3Rs implications as it reduces the number of false-positive founders carried forward for breeding. It also enables researchers to take advantage of mosaic founders that carry multiple genome-editing events, so that fewer founder animals may be required to be produced and screened to analyse equivalent numbers of mutagenesis events, thereby resulting in an overall reduction in the number of animals used.
We offer a comprehensive genome engineering resource from design to model production and validation. You can find out more about how our mouse model generation services can support your work on our Services pages.